Medical Assistant
A system that stores up-to-date information from scientific medical sources and assists in clinical decision-making.
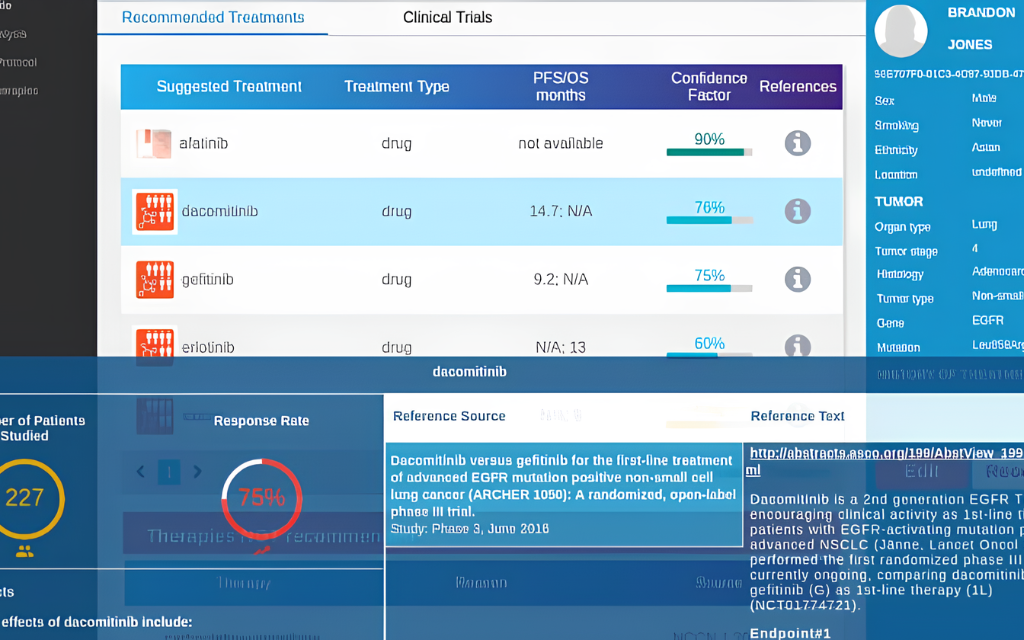
The primary goal of this system is to integrate various medical databases and utilize advanced deep learning algorithms for analyzing and processing information coming from these sources.
Data and Sources
For the development of the system, we used data from several major medical databases such as PubMed, ClinicalTrials.gov, and DrugBank. These resources contain millions of scientific articles, clinical trials, and detailed information on pharmaceutical drugs.
One of the key challenges was that most of this data is presented in unstructured text formats, making automated analysis difficult. The problem was how to extract relevant information about diseases, symptoms, medications, and other medical aspects from the vast amount of scientific publications. To address this, we used classical text processing methods, language models, and Named Entity Recognition (NER).
Neo4j Graph Database
One of the key components of the project’s architecture was the Neo4j graph database. This tool was perfect for modeling relationships between various medical entities such as diseases, symptoms, medications, medical research, and their outcomes.
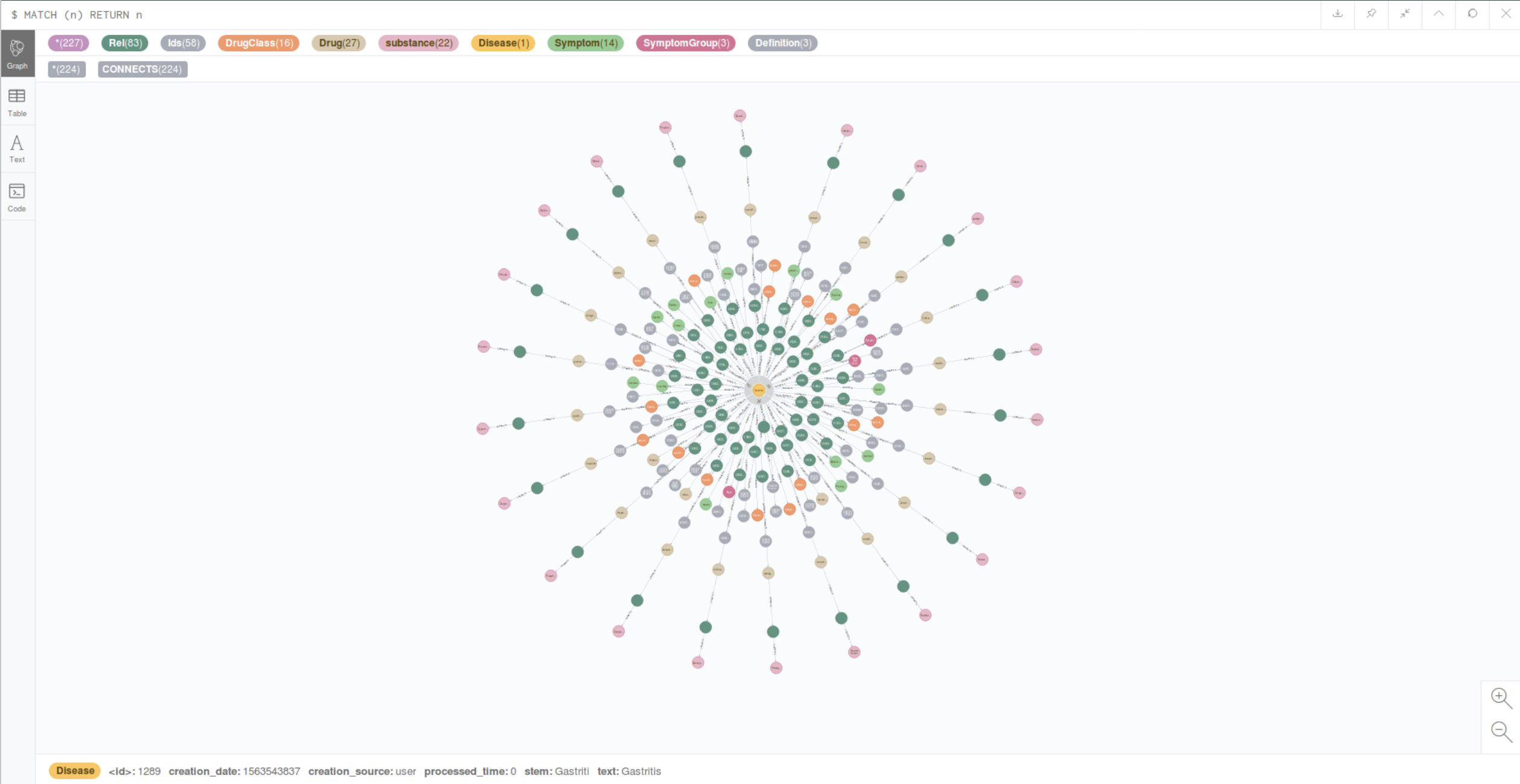
In traditional relational databases, it is difficult to model such complex relationships, whereas a graph model allowed us to represent data as graphs, where nodes represent different entities and edges represent their connections. For example, a disease node could automatically link to a symptom node and a medication node used to treat that disease.
This approach enabled the system to quickly and efficiently explore connections between various medical objects. For instance, if a doctor is interested in a particular disease, the app can show not only a description of the disease but also associated symptoms, treatment methods, and ongoing research related to that disease.
Medical Text Processing and Entity Extraction
Medical publications often contain specialized terminology, which makes them difficult to understand and analyze. The challenge in processing such texts was the need for precise extraction of entities like disease names, symptoms, medications, as well as medical markers such as biomarkers or genetic factors.
To address this, we used our own transformer-based models, specifically trained to work with medical terminology. These models were trained on large volumes of medical texts, enabling them to extract the necessary information effectively. We applied NER techniques to identify and classify entities, as well as algorithms for context analysis, which improved the accuracy of entity extraction, especially for rare diseases or new drugs.
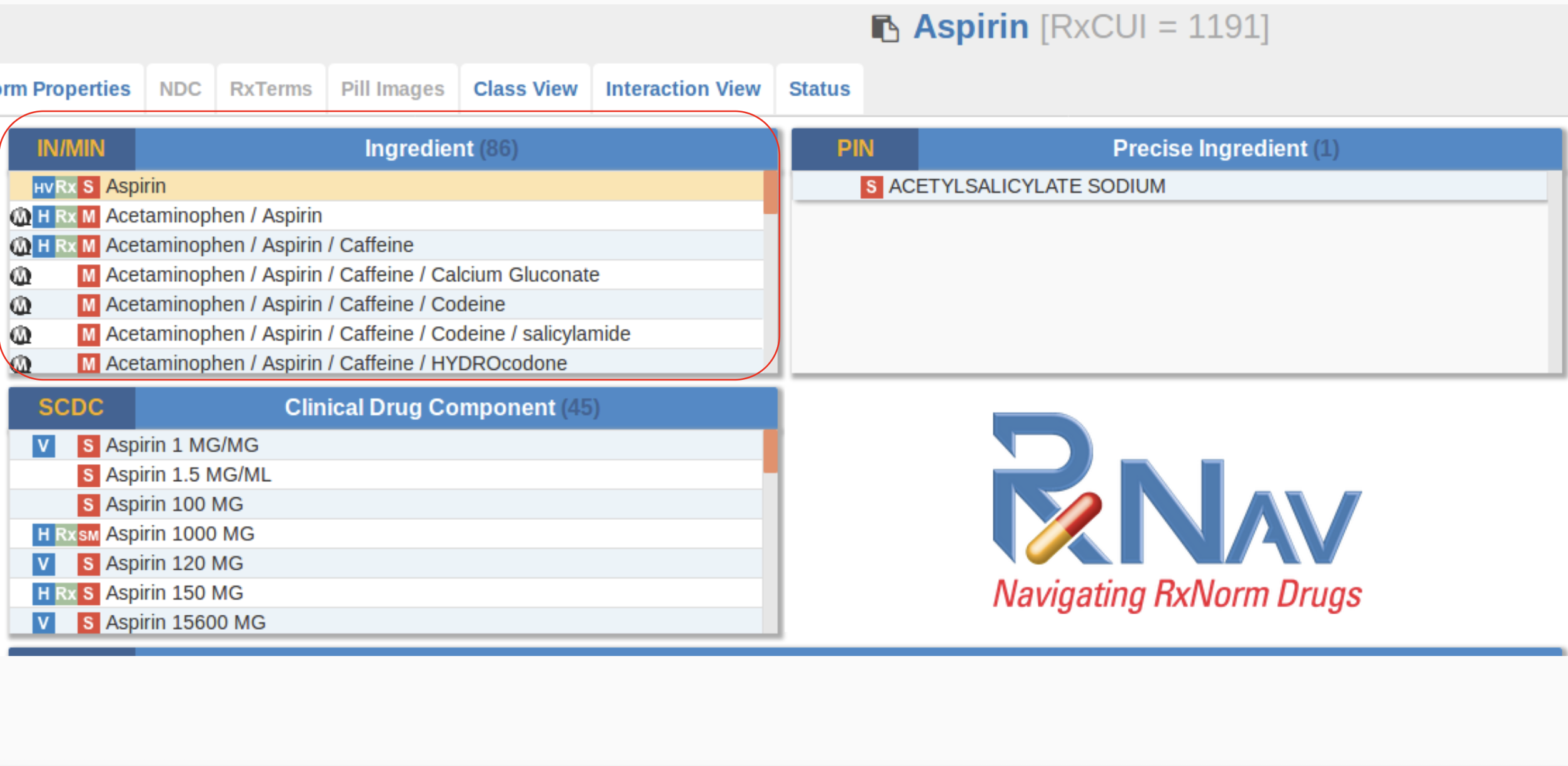
We employed transformer-based models like BERT and its specialized versions for medical texts (something like a custom implementation of BioBERT), which showed excellent results in analyzing medical publications. It was crucial not only to extract entities but also to classify them based on their importance, reliability, and relevance to specific medical practices.
Reliability of Research
One of the most important aspects of the system was classifying scientific research based on its reliability. A doctor needs to rely on verified sources, so it was essential to solve the task of evaluating the reliability and relevance of scientific articles. We achieved this by classifying publications based on a range of factors such as research methodology, author reputation, citation count, and data quality.
For classification, we used classical machine learning methods to automatically assess the credibility and reliability of research based on these factors. This way, the app not only provided data but also added valuable context about the credibility of the information within the current medical landscape.
Data Processing and System Backend
Since the project involved working with large volumes of data, the system’s architecture was designed with scalability and performance in mind. All data was stored in two main types of databases: relational (e.g., PostgreSQL) for structured data and graph databases (e.g., Neo4j) for interconnected medical entities.
The app’s backend was built using a microservice architecture in Python, which provided flexibility and scalability. The main framework for data processing was Django, which was used to create a RESTful API that allowed the system to interact with users. All request processing occurred on the server, where data was extracted from various databases, processed using machine learning and NLP algorithms, and returned as structured responses.
For model training, we used our own computational servers with GPUs (e.g., NVIDIA Tesla V100, A100), which provided the necessary performance for efficiently handling large data volumes and fine-tuning transformer models.
Open Medical Terms Databases
A key challenge in working with medical data was the vast amount of specialized medical terminology used in scientific articles.
We developed a system that efficiently recognized and classified terms related to diseases, symptoms, medications, and other useful and important biomedical entities. To achieve this, we used ontologies like SNOMED CT (a terminology system for representing medical concepts) and ICD-10.
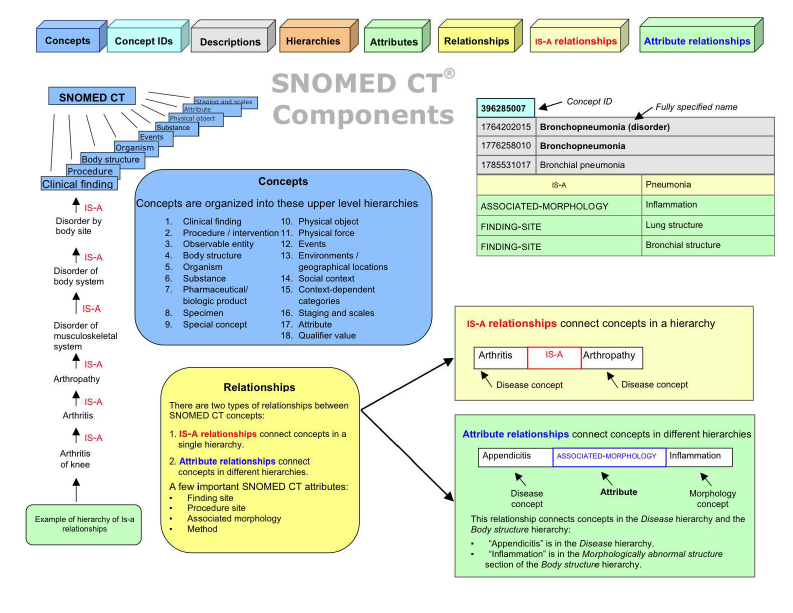
These ontologies allowed us to standardize medical terminology and integrate data from various sources.
As a result, the system could not only extract key medical terms from texts but also correctly classify them, which increased the accuracy and usefulness of the provided data. This is especially important in the medical field, where even the slightest error in interpreting a term can lead to serious consequences.